Science丨施一公组报道酿酒酵母pre-B complex状态剪接体电镜结构
| 导读 | Science在线发表了施一公课题组题为“Structures of the fully assembled Saccharomyces cerevisiae spliceosome before activation”的研究论文,解析了酿酒酵母平均分辨率分别为3.3-4.6 Å以及3.9 Å的催化前复合物B complex状态(pre-B complex )和B complex状态(pre-catalytic spliceosome)的剪接体电镜结构。清华大学白蕊、万蕊雪和闫创业该文的共同第一作者,施一公教授为本文的通讯作者。 |
5月25日凌晨,Science在线发表了施一公课题组题为“Structures of the fully assembled Saccharomyces cerevisiae spliceosome before activation”的研究论文,解析了酿酒酵母平均分辨率分别为3.3-4.6 Å以及3.9 Å的催化前复合物B complex状态(pre-B complex )和B complex状态(pre-catalytic spliceosome)的剪接体电镜结构。清华大学白蕊、万蕊雪和闫创业该文的共同第一作者,施一公教授为本文的通讯作者。

在剪接反应过程中,多种蛋白质-核酸复合物及剪接因子按照高度精确的顺序发生结合和解聚,依次形成预组装复合物U4/U6.U5 tri-snRNP以及至少7个状态的组装剪接体B、Bact、B*、C、C*、P以及ILS复合物。
早在2015年8月,施一公课题组就在Science上背靠背地发表两篇研究长文,报道了裂殖酵母3.6 Å分辨率下的内含子套索剪切体ILS complex的冷冻电镜结构,揭示了剪接体剪接活性位点的原子详情和总的蛋白和RNA的组织形式,并阐释了剪切体对pre-mRNA进行剪接的基本工作原理。此后,施一公教授课题组与MRC分子生物学实验室Nagai研究组、德国马普生物物理化学研究所Reinhard Lüehrmann教授一起互补或背靠背地解析了酿酒酵母或人源的预组装复合物U4/U6.U5 tri-snRNP、催化前复合物B complex、激活状态复合物Bact complex、第一步催化反应复合物C complex、第二步催化激活状态下的C* complex、内含子套索剪接体ILS complex以及剪接体呈现RNA剪接反应完成后状态P complex的三维结构。
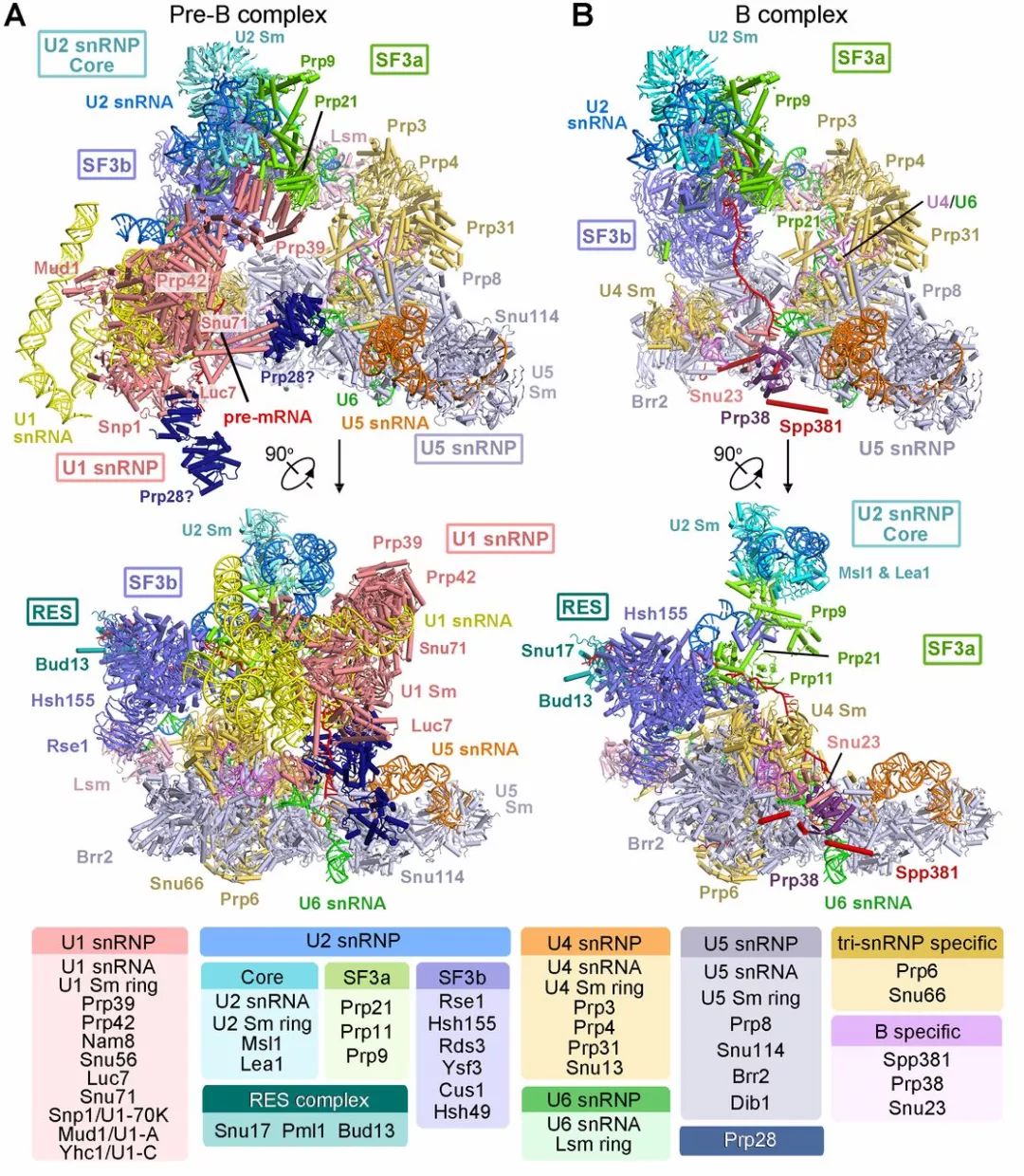
酿酒酵母 pre-B complex 和 B complex状态下剪接体电镜结构
附施一公实验室有关剪接体的论文列表:
1、Yan, C., Hang, J., Wan, R., Huang, M., Wong, C. C., & Shi, Y. (2015). Structure of a yeast spliceosome at 3.6-angstrom resolution. Science, 349(6253), 1182-1191.
2、Hang, J., Wan, R., Yan, C., & Shi, Y. (2015). Structural basis of pre-mRNA splicing. Science, 349(6253), 1191-1198.
3、Wan, R., Yan, C., Bai, R., Wang, L., Huang, M., Wong, C. C., & Shi, Y. (2016). The 3.8 Å structure of the U4/U6. U5 tri-snRNP: Insights into spliceosome assembly and catalysis. Science, 351(6272), 466-475.
4、Yan, C., Wan, R., Bai, R., Huang, G., & Shi, Y. (2016). Structure of a yeast activated spliceosome at 3.5 Å resolution.Science, 353(6302), 904-911.
5、Yan, C., Wan, R., Bai, R., Huang, G., & Shi, Y. (2016). Structure of a yeast step II catalytically activated spliceosome.Science, aak9979.
6、Wan, R., Yan, C., Bai, R., Huang, G., & Shi, Y. (2016). Structure of a yeast catalytic step I spliceosome at 3.4 Å resolution.Science, 353(6302), 895-904.
7、Zhang, X., Yan, C., Hang, J.,Finci, L., Lei, J., & Shi, Y. (2017).An Atomic Structure of the Human Spliceosome.Cell,169, 1–12
8、Wan, R., Yan, C., Bai, R.,Lei, J., & Shi, Y. (2017).Structure of an Intron Lariat Spliceosome from Saccharomyces cerevisiae.Cell,171, 1–13
9、Bai, R., Yan, C., Wan, R., Lei, J., & Shi, Y. (2017). Structure of the Post-catalytic Spliceosome from Saccharomyces cerevisiae. Cell, 171(7), 1589-1598.
10、Zhan, X., Yan, C., Zhang, X., Lei, J., & Shi, Y. (2018). Structure of a human catalytic step I spliceosome. Science, eaar6401.
综述:
Shi, Y. (2017). Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nature Reviews Molecular Cell Biology.
Shi, Y. (2017). The Spliceosome: A Protein-Directed Metalloribozyme. Journal of Molecular Biology, 429(17), 2640-2653.
(转化医学网360zhyx.com)
 腾讯登录
腾讯登录
还没有人评论,赶快抢个沙发